October 2024 - Today
AI Research Engineer @ DentalMonitoring

Supervisors : Guillaume Ghyselinck
As an AI Research Engineer at Dental Monitoring, I Innovate end-to-end projects, from conception to deployment. I design and implement AI-based recognition systems for remote orthodontic monitoring, leveraging advanced machine learning techniques. Proficient in computer vision, biomedical engineering, medical imaging, and general AI research, I collaborate with a dynamic R&D team.
July 2021 - September 2024
PhD Candidate | Machine Learning Engineer @ Philips Health Technology Innovation - AI Research Hub France
 Supervisors : Pr. Loic Boussel, Dr. Nicolas Villain
Supervisors : Pr. Loic Boussel, Dr. Nicolas Villain
I completed a CIFRE PhD in collaboration with Philips Health Technology Innovation, INSA Lyon, and Hospices Civils de Lyon (HCL), under the supervision of Prof. Loïc Boussel, Dr. Nicolas Villain, Dr. Olivier Nempont, and Dr. Alexandre Popoff. My research explored the development of AI-driven pathology detection systems for conventional CT scans, combining computer vision, medical imaging, and deep learning. Working closely with teams in healthcare and research, I contributed to the design of automated tools that could potentially improve the precision and efficiency of clinical workflows. This research led to several publications, and I gained valuable experience in Python and deep learning frameworks throughout the process.
March 2020 - August 2020
Computer Vision and Deep Learning Intern @ GE Healthcare
 Supervisors : Pr. Serge Muller, Dr. Andrei Petrovskii
Supervisors : Pr. Serge Muller, Dr. Andrei Petrovskii
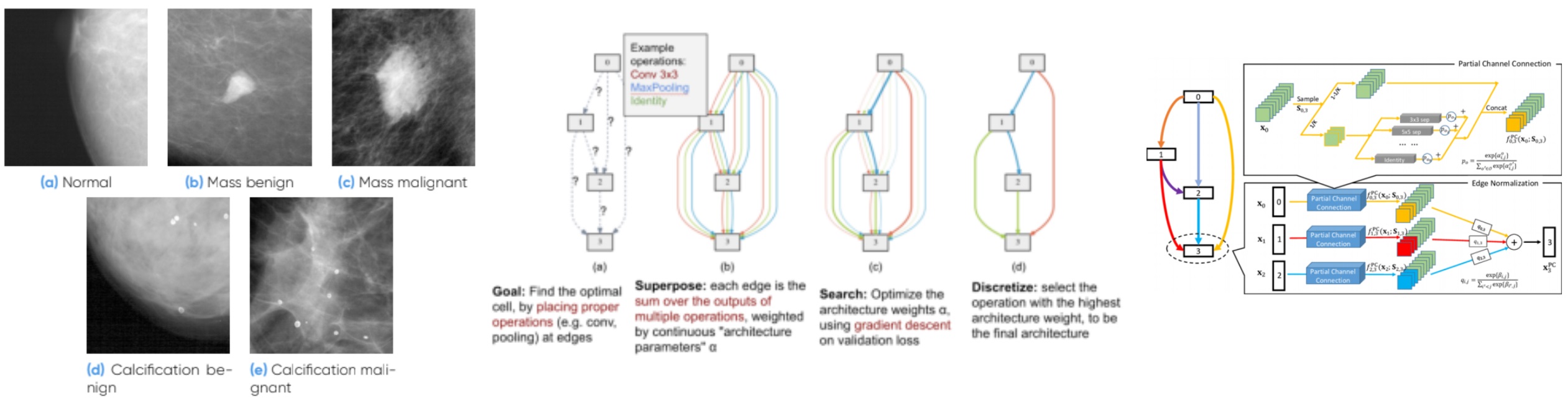
Investigation of automatic search methods for neural network hyperparameters (Neural Architecture Search).
Application to Mammographic Data within WHARe team (Women’s Health Applied Research)
During my internship, I collaborated with experts to advance mammography systems for clinical diagnosis, focusing on the application of deep learning techniques to enhance the accuracy of mammographic data classification. Specifically, we explored the potential of a gradient-based Neural Architecture Search (NAS) method, DARTS (Differentiable Architecture Search), to optimize model performance for classification tasks. Through extensive experimentation, we successfully achieved state-of-the-art results, surpassing existing models in mammography classification. This project not only provided valuable hands-on experience with AI-driven research but also inspired my decision to pursue a PhD in medical imaging and deep learning.
Publications
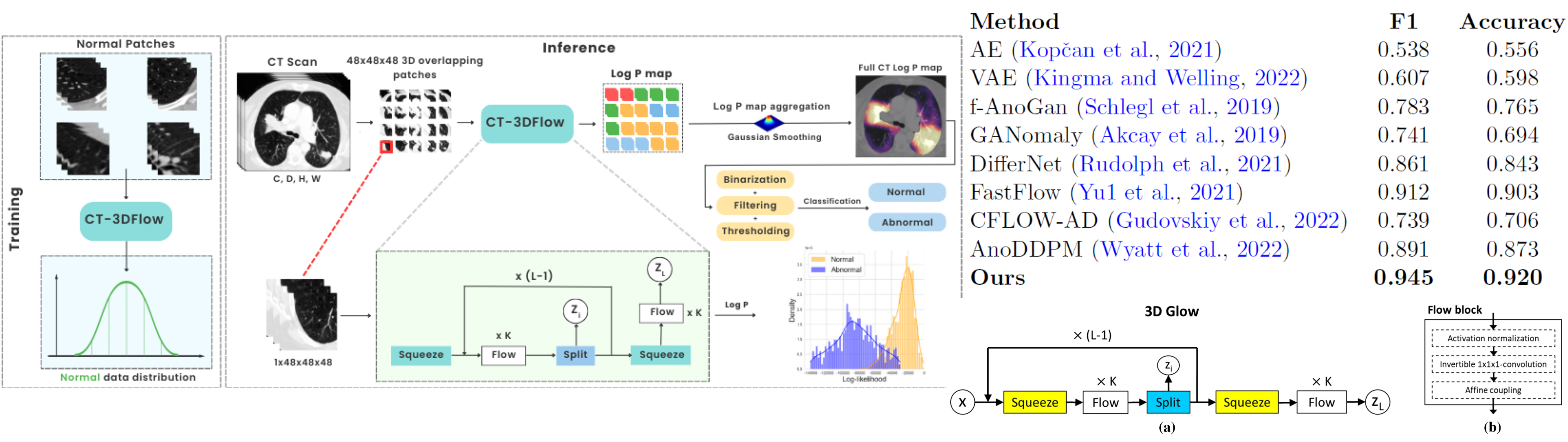
Leveraging Normalizing Flows for Unsupervised Detection of Pathological Pulmonary in CT scans
A. Djahnine, E. Jupin-delevaux, O. Nempont, S. A. Si-Mohamed, V. Cottin, A. Popoff, P. Douek, L. Boussel
IEEE TRANSACTIONS ON MEDICAL IMAGING - IEEE TMI[In revision]
Weakly-Supervised learning based Pathology Detection and Localization in 3D Chest Scans
A. Djahnine, E. Jupin-delevaux, O. Nempont, F. Craighero, S. A. Si-Mohamed, V. Cottin, P. Douek, A. Popoff, L. Boussel
The International Journal of Medical Physics Research and Practice (Medical Physics)
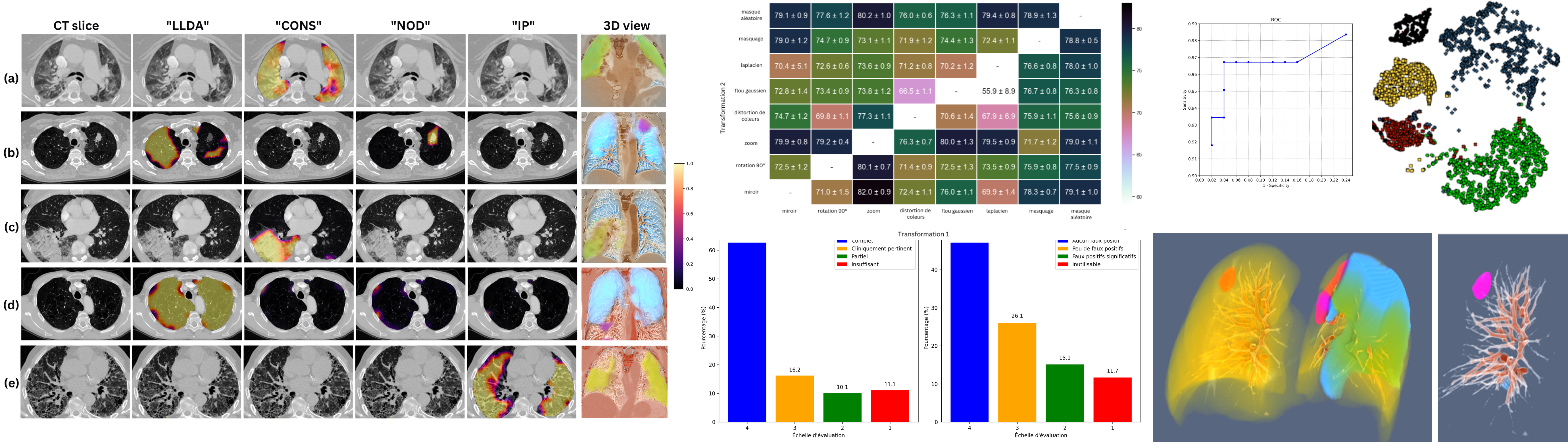
Detection and severity quantification of pulmonary embolism with 3D CT data using an automated deep learning-based artificial solution
A. Djahnine, C. Lazarus, M. Lederlin, S. Mulé, R. Wiemker, S. Si-Mohamed, E. Jupin-Delevaux, O. Nempont, Y. Skandarani, M. De Craene, S. Goubalan, C. Raynaud, Y. Belkouchi, A. Ben Afia, C. Fabre, G. Ferretti, C. De Margerie, P. Berge, R. Liberge, N. Elbaz, M. Blain, P. Brillet, G. Chassagnon, F. Cadour, C. Caramella, M. El Hajjam, S. Boussouar, J. Hadchiti, X. Fablet, A. Khalil, H. Talbot, A. Luciani, N. Lassau and L. Boussel
Diagnostic and Interventional Imaging Journal (DII)
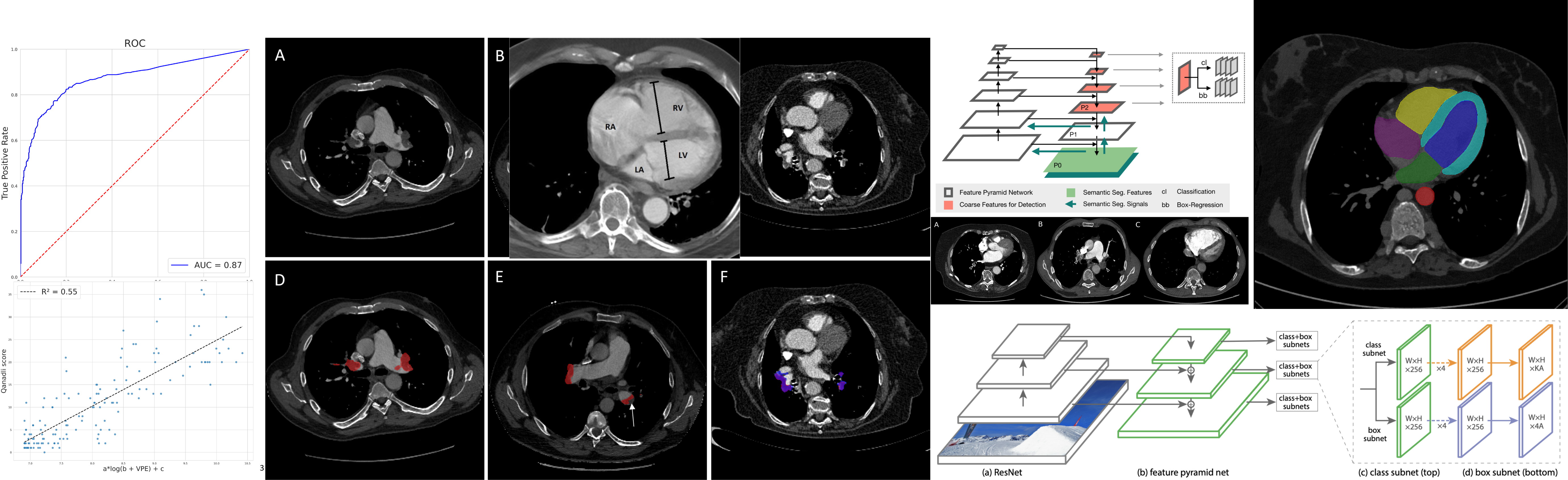
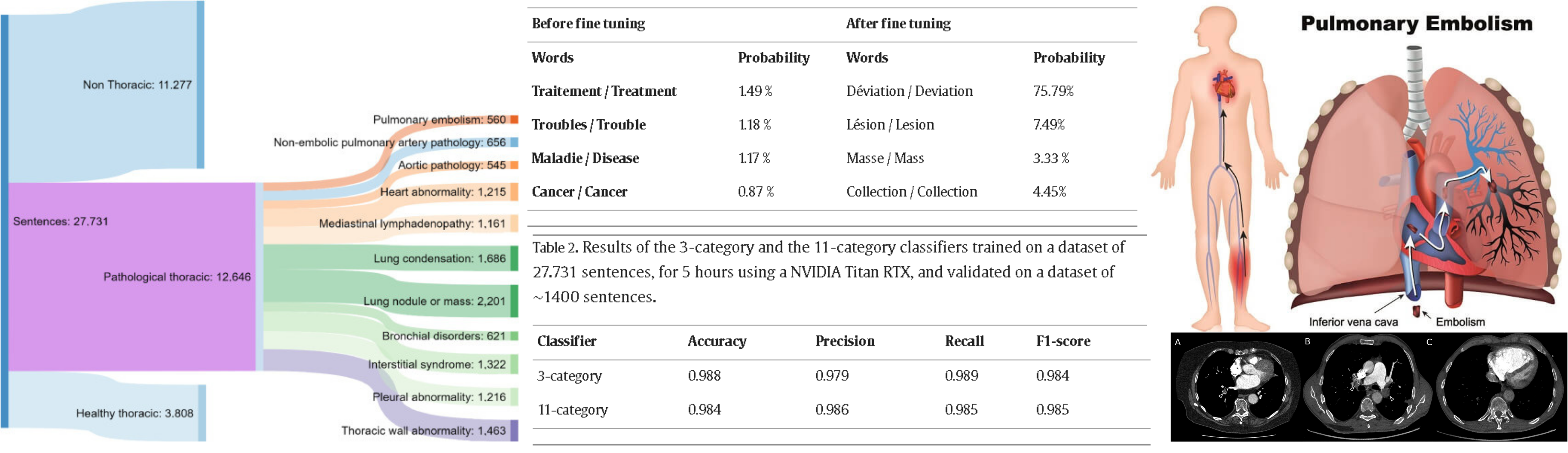
BERT-based natural language processing analysis of French CT reports: Application to the measurement of the positivity rate for pulmonary embolism
E. Jupin-Delevaux, A. Djahnine, F. Talbot, A. Richard, S. Gouttard, A. Mansuy, P. Douek, S. A. Si-Mohamed, L. Boussel
Research in Diagnostic and Interventional Imaging Journal (ReDII)
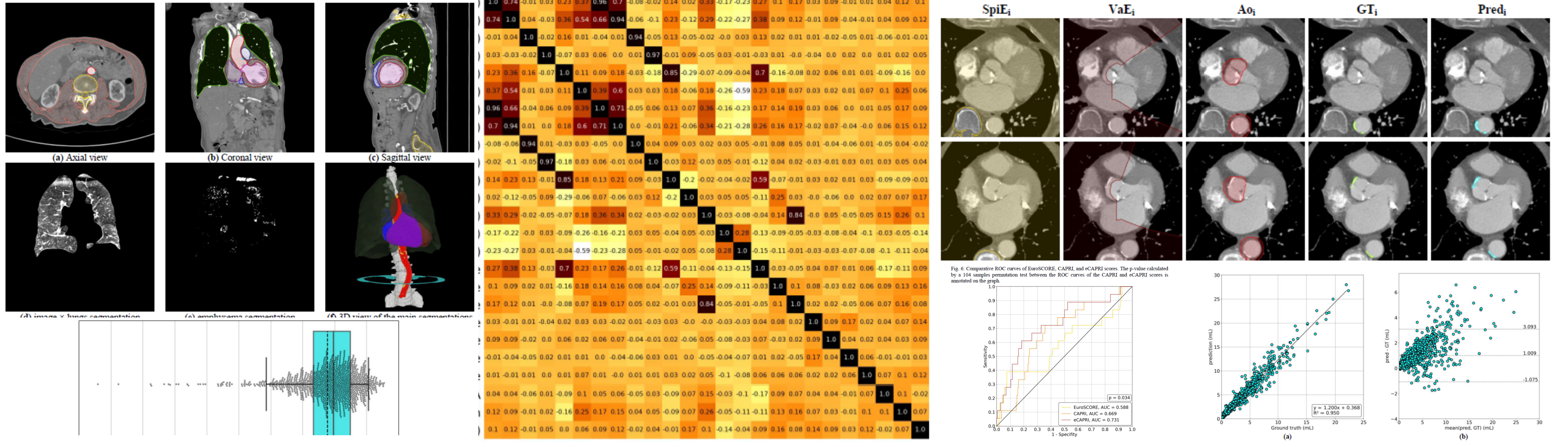
Automatization and development of the TAVI CAPRI risk score with imaging biomarkers
P.-J. Lartaud, B. Harbaoui, A. Djahnine, O. Nempont, J.-M. Rouet, Anna Vlachomitrou, B. Riche, M. Rabilloud, S. Si-Mohamed P. Douek, F. Derimay, G. Rioufol, P. Lantelme, L. Boussel
Radiology Advances : Oxford AcademicIn revision
Leveraging 3D Normalizing Flows for Unsupervised Detection of Pathological Pulmonary CT Scans
Djahnine Aissam, Popoff Alexandre, Jupin-Delevaux Emilien, Cottin Vincent, Nempont Olivier, Boussel Loic
Arxiv
Conferences & Summer Schools
16 th IEEE International Conference on Signal Processing (ICSP)
21 - 24 October 2022
Beijing, China
Tailored 3D CT contrastive pretraining to improve pulmonary pathology classification
A. Djahnine, A. Popoff, E. Jupin-Delevaux, V. Cottin, O. Nempont, L. Boussel
Oxford Machine Learning Summer School (OxML 2023)(ML x Health Track, In Person)
Certificate of Participation
13 - 16 July 2023
Oxford, United Kingdom
Honors & awards
JFR 2023 Data Challenge WINNER : Pancreatic masses detection in 3D CT scans
- I was a member of the Philips team in collaboration with Hospices Civils de Lyon that won the JFR (les Journées Francophones de Radiologie) data challenge. The solution used deep learning-based algorithm to detect pancreatic masses in 3D CT scans.

JFR 2022 Data Challenge WINNER : Pulmonary embolism detection in 3D CT scans
- I was a member of the Philips team in collaboration with Hospices Civils de Lyon that won the JFR (les Journées Francophones de Radiologie) data challenge. The solution used Deep Learning for Computer Vision to detect pulmonary embolism in CT scans. ( Challenge Paper, Our Solution (Journal Paper))

Academic Services
Reviewer : 20th IEEE International Symposium on Biomedical Imaging, ISBI 2023
Academic Tutor : Assisted undergraduate students with academic projects and lab sessions, focusing on mathematics, linear algebra problem-solving, analog and digital electronics applications, as well as programming in C, MATLAB, and VHDL at Sorbonne University (2019)
Research Projects
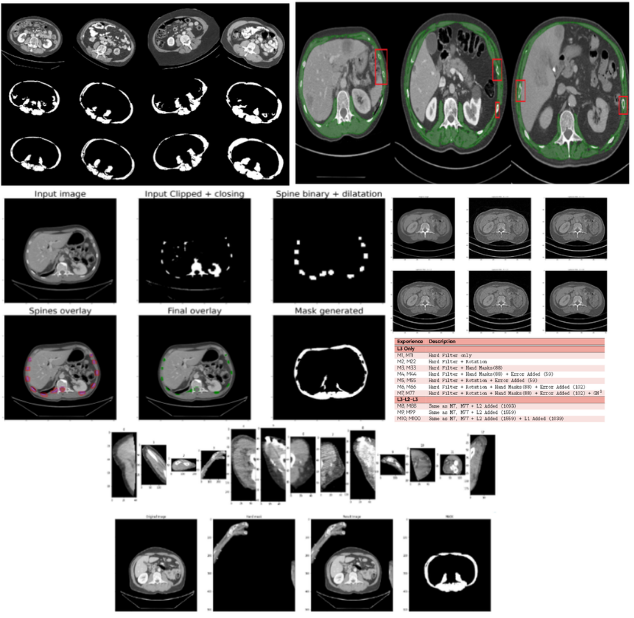
PRE-PHD SHORT CONTRACT WITH HOSPICES CIVILS DE LYON
Segmentation and measurement of skeletal muscle areas on CT scans
- This project aims to segment and measure skeletal muscle areas on CT scans, specifically at the L3, L2, and L1 vertebrae levels, to improve sarcopenia estimation using deep learning-based methods. By leveraging advanced data augmentation techniques, such as applying filters, rotations, and adding artificial elements, the dataset’s diversity and robustness were significantly improved. Post-processing methods were developed to remove artifacts like ribs, ensuring cleaner and more accurate segmentations. The approach showed promising results in accurately segmenting muscle areas, leading to more reliable sarcopenia assessments and potential further improvements.
Computer Vision and Deep Learning Intern at GE Healthcare
Investigation of automatic search methods for neural network hyperparameters (Neural Architecture Search). Application to Mammographic Data within WHARe team (Women’s Health Applied Research)

- In this internship, we focused on improving mammography systems for clinical diagnosis by utilizing deep learning techniques. We specifically explored the efficiency of a gradient-based NAS method called DARTS (Differentiable Architecture Search) for classification tasks on mammographic data. Through experiments, we achieved state-of-the-art results on mammography data classification, outperforming existing models.
Unsupervised Spatiotemporal Data Inpainting
- This a PyTorch implementaiton of this paper : Unsupervised Spatiotemporal Data Inpainting Under review at ICLR 2020.
- The code is implemented in collaboration of : Ilyas aroui Github, Raouf Keskes Github
- Inpainting spatio-temporal sequences is an active research topic that relies heavily on supervision with large datasets. In this work, we consider the problem of reconstructing missing information with an unsupervised learning approach. Following the work of Kim et al. We train a generative model on the occluded sequences. We ensure that the models captured both frame-based and sequential based information. Our proposed model is adapted to large-scale images and can be used to different types of sequences and occlusion processes.
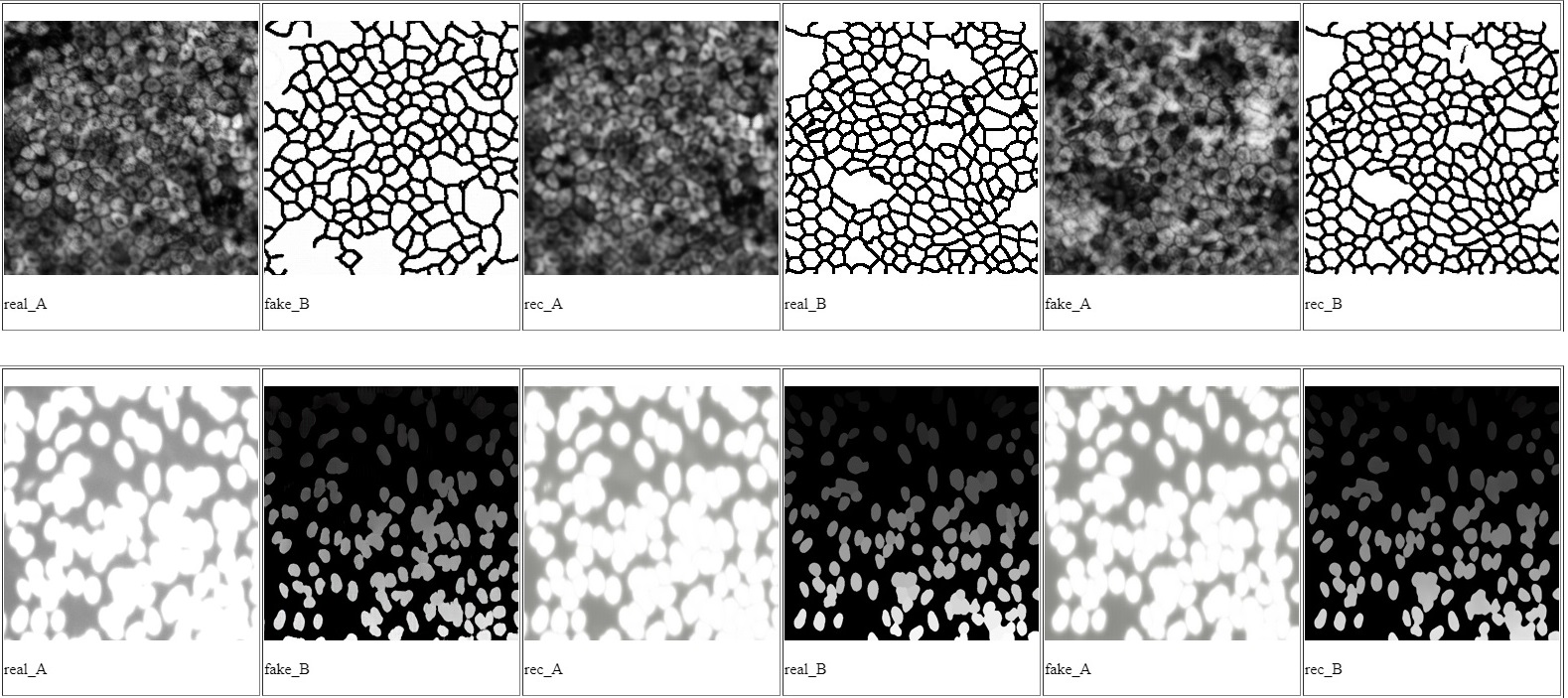
Cell Images Segmentation Using Cycle Generative Adversarial Network

- During our 6-month curriculum project with Institut Pasteur and Sorbonne University, we focused on cell image segmentation for various medical applications. Recognizing the significance of labeled data for training accurate convolutional neural networks (CNNs), we adopted a cycleGAN framework to overcome the challenge of limited data. Our research objectives were two-fold: first, to generate synthetic cell images that mimic the distribution of input images for data augmentation, combining them with real cell images to train a context-aware CNN for precise cell segmentation. Second, we proposed a segmentation method based on cycle-consistent generative adversarial networks (CycleGANs), which allowed us to train the model even in the absence of prepared image-mask pairs.
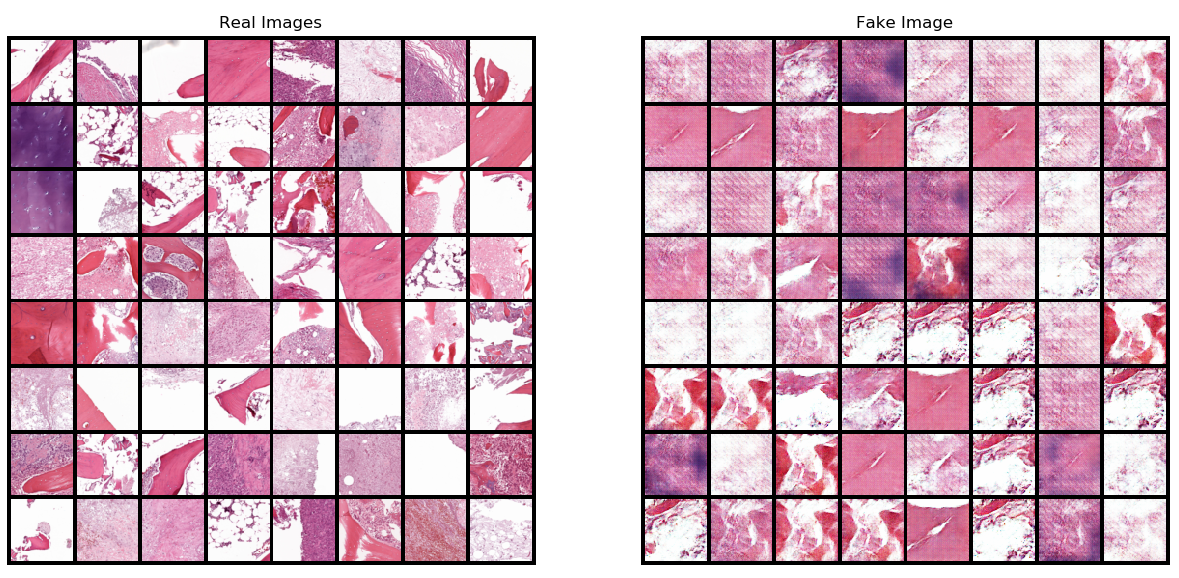
Histopathological Images Generation Using Generative Adversarial Network
- Here, we implement an approach to histopathological image generation that overcomes the challenge of dataset size (small datasets) in medical field by utilizing a GAN framework (Generative adversarial network). The main objective of this research project is :
- Generate synthetic histopathological images that model the distribution of the input images for data augmentation. Use both of the synthetic and real images for training in different tasks (detection/segmentation/tracking).
Online Courses
- Coursera : Deep learning Specialization
- Udemy :
- Python for Data Science and Machine Learning Bootcamp Certificate.